In synthetic biology and metabolic engineering, there is a strong demand for orthogonal or externally controlled regulation of gene expression. RNA-based regulatory devices like riboswitches represent a promising toolbox.
Riboswitch design to regulate gene expression
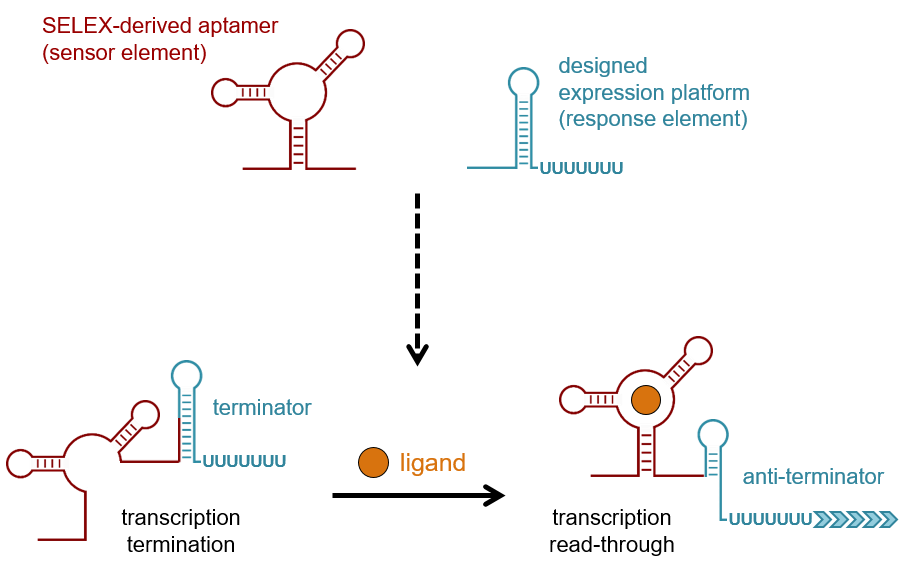
These regulatory elements are usually embedded in the 5′-untranslated region of the mRNA. They allow for a fast and direct control of gene expression, as no synthesis of regulatory proteins is required. Riboswitches show a modular composition of an aptamer domain as sensor and an expression platform as response element. The aptamer specifically interacts with a ligand, modulating the secondary structure of the adjacent response element that controls the expression of the downstream located genes, in most cases at the level of transcription or translation. The two domains allow a free combination of different modules in a plug-and-play-like mode for synthetic devices designed analogously to natural examples.
In silico predicted artificial riboswitches control gene expression in vivo
Synthetic riboswitches are often based on in vitro selected aptamers in combination with natural expression platforms. One focus of our lab and our collaboration partners is the understanding and usage of in silico predicted expression platforms. We constructed the first synthetic riboswitches that regulate the transcription of a target gene in a ligand-dependent way. Furthermore, we use our design pipeline to construct riboswitches that control the processing of tRNA precursors – a promising strategy that adds a new layer of control in tRNA maturation and that could be used as a regulatory tool in tRNA-dependent genetic code expansion.